Extract Genomic Sequences
To better help biologists accessing genomics data, we integrated Sequence Extractor tool, located on SeqExtractor tab under Tools category.
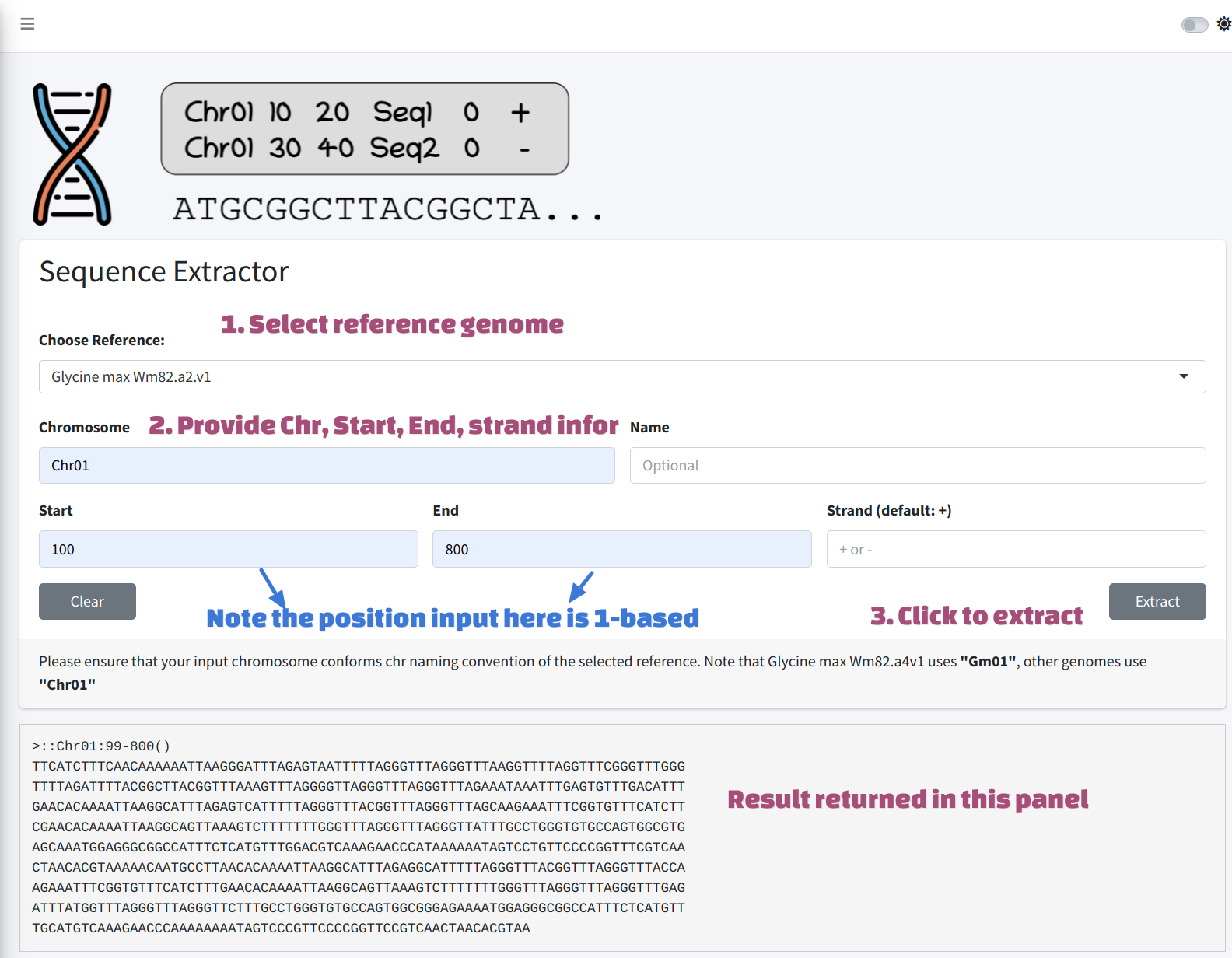
Extract on the fly
User could select reference genome from the dropdown menu, and then provide a valid genomic coordinates including chromosome, start, end and strand, click Extract button, the result will be returned in the lower panel.
The coordinates here is 1-based.
Please also comply the naming convention of the reference genome chromosomes.
The chromosome names of Glycine max Williams 82 a4v1 use Gm as prefix, e.g. “Gm01”, while other genomes use Chr as prefix.
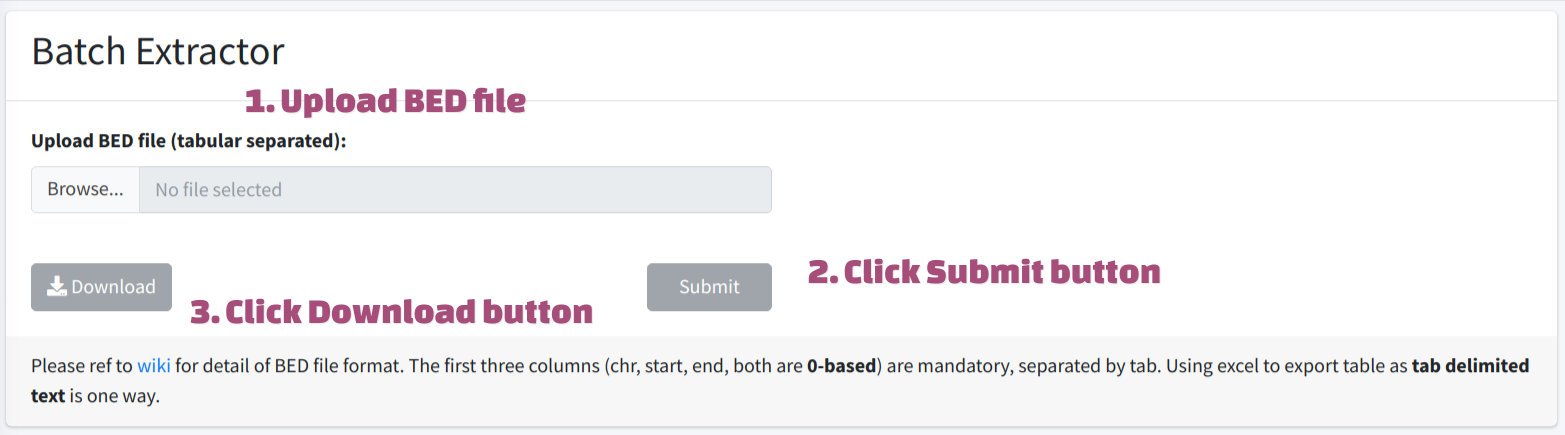
Batch Extracting
Batch extracting could also be achieved. User have to firstly choose reference genome from the drop-down menu mentioned above, then provide a valid BED file (max 500 rows) to the server. Please refer to BED wiki to ensure the format is correct. The first three columns (chr, start, end, both are 0-based) are mandatory, separated by tab. The fourth column indicates strand information, which is optional, server will extract sequence from plus strand by default. To generate BED input, user could use excel to export table as tab delimited text file.
The coordinates in BED file is 0-based.
The Submit button will be enabled when uploading finished. User could click the button to submit the job.
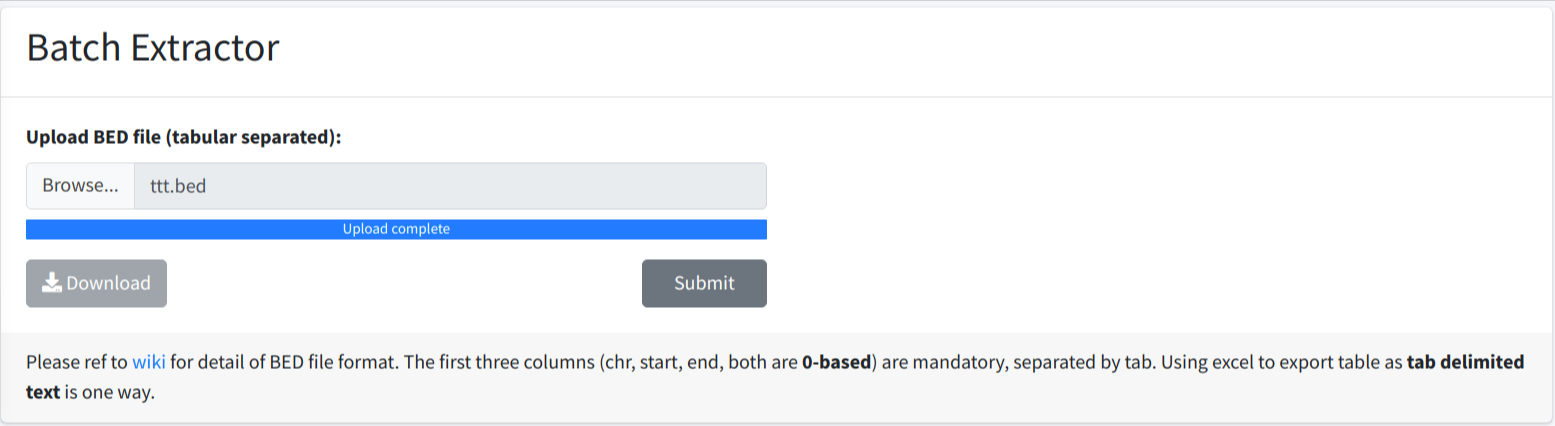
A notice will popup when extracting process is done, user could then click Download button to download the result.

