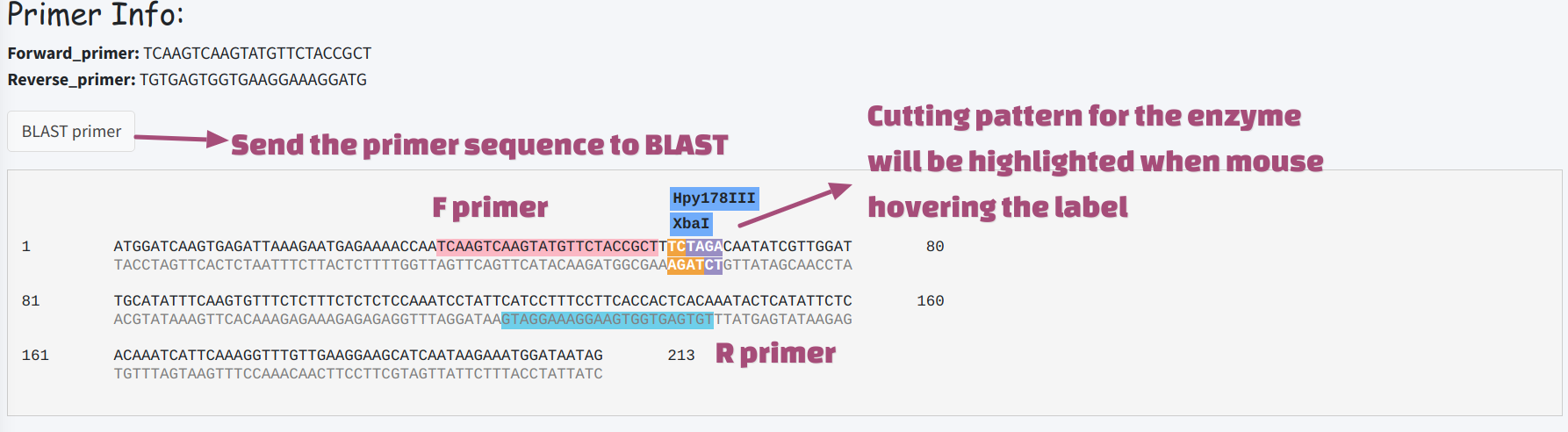
Design Primer
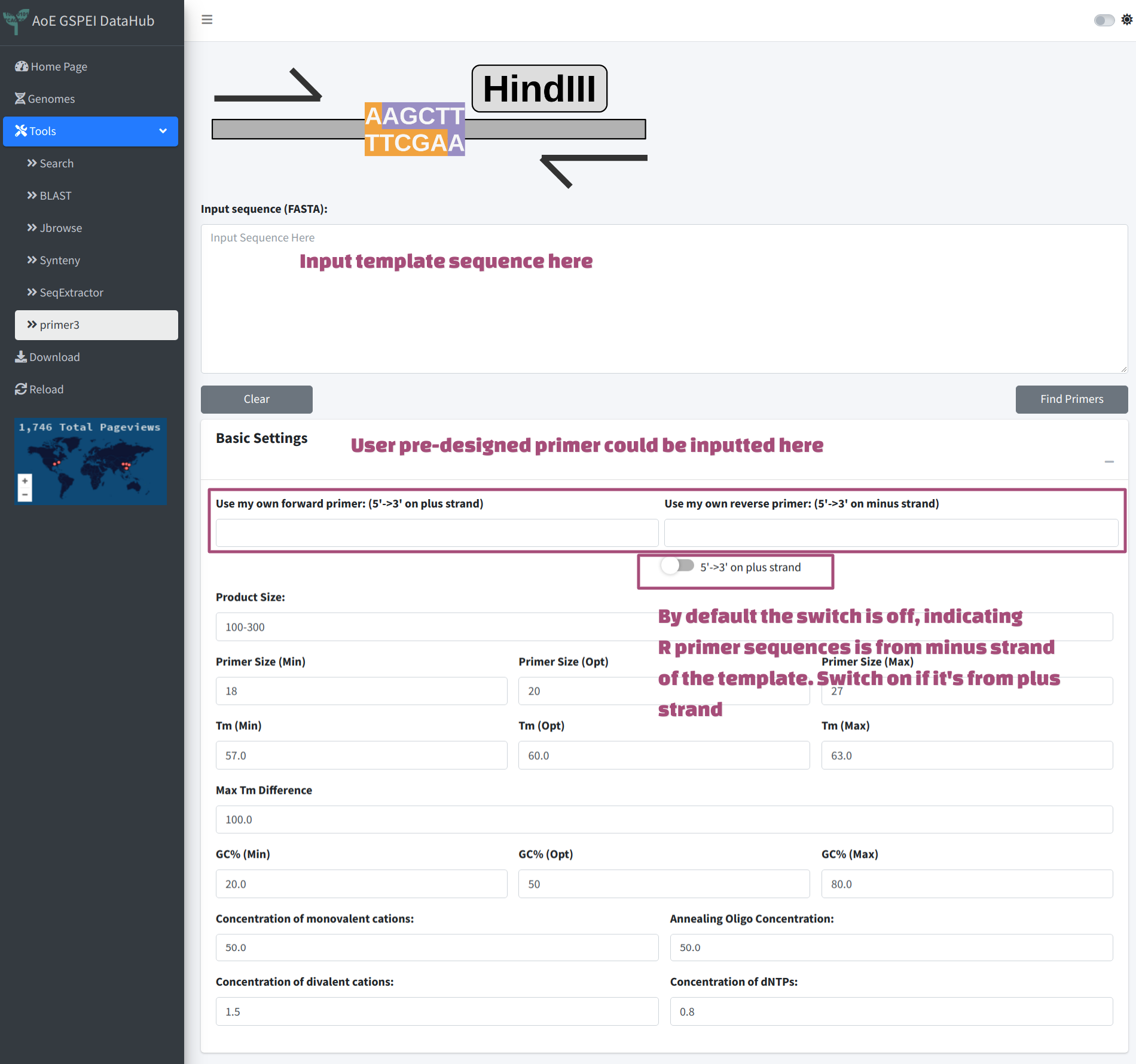
Primer Design function was located on Primer3 tab under Tools category. This module uses primer3 to provide an universal primer discover function for any user submitted sequence or sequences from gene search result. Restriction Enzyme sites in the product will be predicted based on REBASE (20210430) and illustrated in the result panel.
Optionally, user could provide their pre-designed primer sequences as input, and evaluate the primer according to primer3’s report.
The primer sequences could also be sent to BLAST tab and searched against target
transcriptome or genome database. BLAST parameters will be adjusted for short query
sequence (blastn-short), and user could check the specificity of the primer based
on BLAST result.
The 10 best results will be reported in a summary table, with the information of primer sequences, position, length, annealing temperature, GC content, as well as the product length.
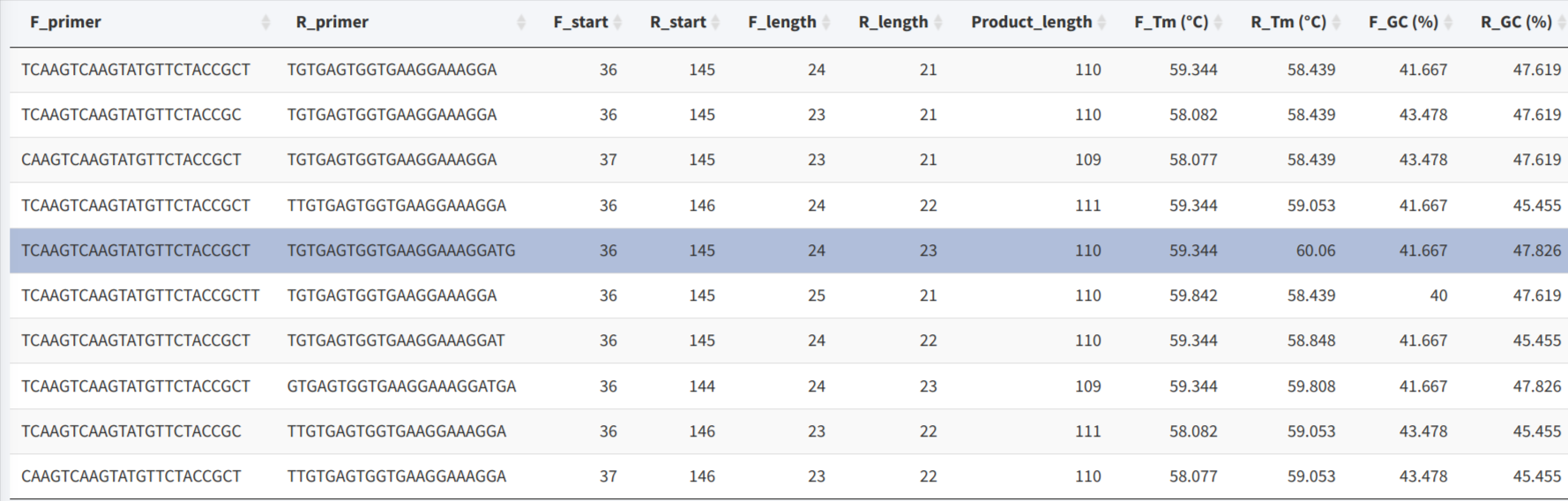
Detail information will be shown after the record be chosen in the table.